As my project takes a design approach to engage with a proposed second phase of the Luisi’s lab random synthesisation of proteins, it has been necessary for me to continue to focus on the technical process of doing so. While the process I have described so far is the initial step in synthesising a protein, it is essential for me to understand regardless of the specific techniques we use to attempt to produce a novel protein. Moreover, understanding and participating in synthesising a protein is a necessary step in my project as it’s difficult -if not impossible -to collaboratively engage in the difference between randomisation and design techniques through actually synthesising a protein without understanding, and engaging in the experimental processed through which it is expressed. As such, I am continuing to focus on the laboratory process of protein synthesis in which I am participating, while opening up our technical-practical the discussion to questions of design.
As our first attempt at inserting a DNA fragment into a plamid (which I should have explained in my first blog is an independent DNA molecule found in bacterial cells) was unsuccessful we repeated the process, with some changes.
The reason why the fragment did not successfully insert Cristiano surmised was because the plasmid we had cut so we could insert the DNA had rejoined (self-ligased) without the insert. To prevent this occurring again, we added an enzyme into the solution we prepared for cutting another plasmid that would inhibit the plasmid from rejoining. We then ran a test to check whether on this occassion we had been successful. However, the test indicated that in all likelihood the fragment had only been attached to one end of the plasmid, forming a single length rather than a modified circle of plasmid DNA.
Despite out uncertain result Critiano decided to continue the cloning process and insert the plasmid into a bacterial host as the test is not always sensitive enough to register a positive result, and it is a relatively simple procedure to do. By attempting to insert our modified plasmid into a bacterial cell (ecoli to be exact) if we had actually been successful in doing so, we hoped to isolate it from the other possible products that are produed when cloning DNA. Rather than running a battery of tests to verify whether we had achieved our aim, on this occassion after isolating the plasmid DNA from the cells we sent it to a company for sequencing.
Unfortunately, once again, the test came back negative. (I’m told this is common refrain in bioscience.) The sequence returned to us was that of our original, or wild-type plasmid as it is called when being modified (regardless of its actual biotechnological heritage). The results of the test are visualised as a chromatogram, which displays the genetic sequence as a colour coded graph. I’ve included part of it below.
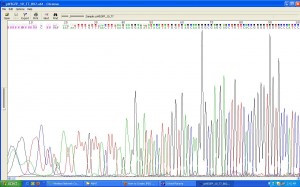
In brief, to visualise the DNA sequence dNTP’s (the separate bases, GATC, comprising DNA) modified with chromophores, (a chemical group capable of light aborption/refection within the visible spectrum) corresponding to their particular code of either GATC are added to the reaction. Then, when the DNA sequences is read each base is visible according to its assigned colour. The test also supplies the code in textual form and the corresponding amino acid sequence for which it codes.
The reason why we got a negative result Cristiano thinks is because the fragment did not have a long enough tail to which the enzyme could attach so that it could cut it, making the sticky-ends necessary to insert it into the plasmid. His interpretation fits with his reading of the test that the fragment may have only been bound to one end of the cut plasmid; Possibly, only one end of the fragment was made sticky when we prepared it for insertion. To rectify the problem, he will “design” new primers with longer tails so that the enzyme that cuts the ends of the fragment are able to attach.

